ORM

Object-Relational Model
The Low-Level API is an object-relational model for machine learning. Each class in the ORM maps to a table in a SQLite database that serves as a machine learning metastore.
The real power lies in the relationships between these objects (e.g. Label→Splitset←Feature and Queue→Job→Predictor→Prediction), which enable us to construct rule-base protocols for various types of data and analysis.
Goobye, X_train, y_test. Hello, object-oriented machine learning.
from aiqc.orm import *
Automatic ‘id’ method argument
If an ORM-based classes is instantiated, then any method called by the resulting object will automatically pass in the object’s self.id in as its first positional argument:
queue = Queue.get_by_id(id)
queue.run_jobs()
However, if the class has not been instantiated, then the id is required:
Queue.run_jobs(id)
Although I did not design this pattern, if you think about it, it makes sense. ORMs allow you to fluidly traverse relational objects. If you had to check the ``object.id`` of everything you returned before interacting with it, then that would ruin the user-friendly experience.
0. BaseModel
The BaseModel class applies to all tables in the ORM. It’s metadata in the truest sense of the word.
Localized timestamps are handled by utils.config.timezone_now(). They are made human-readable via strftime('%Y%b%d_%H:%M:%S') → “2022Jun23_07:13:14”
0a. Methods
└── created_at()
Returns the creation timestamp in human-readable format.
└── updated_at()
Returns timestamp of the most recent update in human-readable format.
└── flip_star()
A way to toggle (favorite/ unfavorite) the is_starred attribute in order to make entries easy to find.
└── set_info()
Add descriptive information about an entry so that you remember why you created it
set_info(name, description)
Argument |
Type |
Default |
Description |
|---|---|---|---|
name |
str |
None |
Short name to remember this entry by |
description |
str |
None |
What is unique about this entry? |
0b. Attributes
Attribute |
Type |
Description |
|---|---|---|
id |
AutoField |
Auto-incrementing integer (1-based, not zero-based) PrimaryKey |
time_created |
DateTimeField |
Records a timestamp when the record is created |
time_updated |
DateTimeField |
Records a timestamp when the record is created. Overwritten every time the record is updated. |
is_starred |
BooleanField |
Used to indicated that the entry is a favorite |
name |
CharField |
Short name to remember this entry by |
description |
CharField |
What is unique about this entry? |
1. Dataset
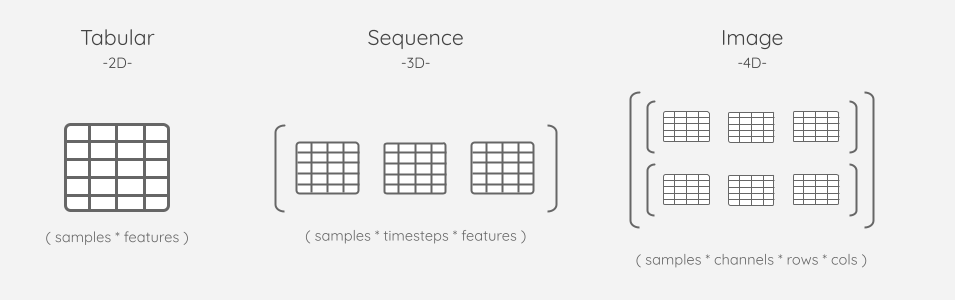
The Dataset class provides the following subclasses for working with different types of data:
Type |
Dimensionality |
Supported Formats |
Format (if ingested) |
|---|---|---|---|
Tabular |
2D |
Files (Parquet, CSV, TSV) / Pandas DataFrame (in-memory) |
Parquet |
Sequence |
3D |
NumPy (in-memory ndarray, npy file) |
npy |
Image |
4D |
NumPy (in-memory ndarray, npy file) / Pillow-supported formats |
npy |
The names are merely suggestive, as the primary purpose of these subclasses is to provide a way to register data of known dimensionality. For example, a practitioner could ingest many uni-channel/ grayscale images as a 3D Sequence Dataset instead of a multi-channel 4D Image Dataset.
Why not 2D NumPy? The
Dataset.Tabularclass is intended for strict, column-specific dtypes and Parquet persistence upon ingestion. In practice, this conflicted too often with NumPy’s array-wide dtyping. We use the best tools for the job (df/pq for 2D) and (array/npy for ND).
1a. Methods
1ai. Registration
Most of the Dataset registration methods share these arguments/ concepts:
Argument |
Description |
|---|---|
ingest |
Determines if raw data is either stored directly inside the metastore or remains on disk to be accessed via path/url. In-memory data like DataFrames and ndarrays must be ingested. Whereas file-based data like Parquet, NPY, Image folders/urls may remain remote. Regardless of whether or not the raw data is ingested, metadata is always derived from it by parsing: 2D via DataFrame and N-D via ndarray. |
rename_columns |
Useful for assigning column
names to arrays or
delimited files that would
otherwise be unnamed.
|
retype |
Change the dtype of data
using
np.types.
All Dataset subclasses
support mass typing via
|
description |
What information does this dataset contain? What is unique about this dataset/ version – did you edit the raw data, add rows, or change column names/ dtypes? |
name |
Triggers dataset
versioning. Datasets that
share a name will be
assigned an
auto-incrementing
|
Ingestion provides the following benefits, especially for entry-level users:
Persist in-memory datasets (Pandas DataFrames, NumPy ndarrays).
Keeps data coupled with the experiment in the portable SQLite file.
Provides a more immutable and out-of-the-way storage location in comparison to a laptop file system.
Encourages preserving tabular dtypes with the ecosystem-friendly Parquet format.
Why would I avoid ingestion?
Happy with where the original data lives: e.g. S3 bucket.
Don’t want to duplicate the data.
sha256? – It’s the one-way hash algorithm that GitHub aspires to upgrade to. AIQC runs it on compressed data because it’s easier and probably less-error prone than intercepting the bytes of the fastparquet intermediary tables before appending the Parquet magic bytes.
Is SQLite a legitimate datastore? – In many cases, SQLite queries are faster than accessing data via a filesystem. It’s a stable, 22 year-old technology that serves as the default database for iOS e.g. Apple Photos. AIQC uses it store raw data in byte format as a BlobField. I’ve stored tens-of-thousands of files in it over several years and never experienced corruption. Keep in mind that AWS S3 is blob store, and the Microsoft equivalent service is literally called Azure Blob Storage. The max size of a BlobField is 2GB, so ~20GB after compression. Either way, the goal of machine learning isn’t to record the entire population within the weights of a neural network, it’s to find subsets that are representative of the broader population.
1ai1. Dataset.Tabular
Here are some of the ways practitioners can use this 2D structure:
Multiple subjects (1 row per sample) * Multi-variate 1D (1 col per attribute) |
Single subject (1 row per timestamp) * Multi-variate 1D (1 col per attribute) |
Multiple subjects (1 row per timestamp) * Uni-variate 0D (1 col per sample) |
Tabular datasets may contain both features and labels
└── Dataset.Tabular.from_df()
dataset = Dataset.Tabular.from_df(
dataframe
, rename_columns
, retype
, description
, name
)
Argument |
Type |
Default |
Description |
|---|---|---|---|
df |
DataFrame |
Required |
pd.DataFrame with int-based single index. DataFrames are always ingested. |
rename_columns |
list[str] |
None |
See Registration |
retype |
np.type / dict(column:np.type) |
None |
See Registration |
description |
str |
None |
See Registration |
name |
str |
None |
See Registration |
└── Dataset.Tabular.from_path()
Dataset.Tabular.from_path(
file_path
, ingest
, rename_columns
, retype
, header
, description
, name
)
Argument |
Type |
Default |
Description |
|---|---|---|---|
file_path |
str |
Required |
Parsed based on how the file name ends (.parquet, .tsv, .csv) |
ingest |
bool |
True |
See Registration. Defaults to True because I don’t want to rely on CSV files as a source of truth for dtypes, and compression works great in Parquet. |
rename_columns |
list[str] |
None |
See Registration |
retype |
np.type / dict(column:np.type) |
None |
See Registration |
header |
object |
None |
See Registration |
description |
str |
None |
See Registration |
name |
str |
None |
See Registration |
1ai2. Dataset.Sequence
Here are some of the ways practitioners can use this 3D structure:
Single subject (1 patient) * Multiple 2D sequences |
Multiple subjects * Single 2D sequence |
Sequence datasets are somewhat multi-modal in that, in order to perform supervised learning on them, they must eventually be paired with a
Dataset.Tabularthat acts as itsLabel.
└── Dataset.Sequence.from_numpy()
Dataset.Sequence.from_numpy(
arr3D_or_npyPath
, ingest
, rename_columns
, retype
, description
, name
)
Argument |
Type |
Default |
Description |
|---|---|---|---|
arr3D_or_npyPath |
object / str |
Required |
|
ingest |
bool |
None |
See Registration. If left blank, ndarrays will be ingested and npy will not. Errors if ndarray and False. |
rename_columns |
list[str] |
None |
See Registration |
retype |
np.type / dict(column:np.type) |
None |
See Registration |
description |
str |
None |
See Registration |
name |
str |
None |
See Registration |
1ai3. Dataset.Image
Here are some of the ways you can practitioners this 4D structure:
Single subject (1 patient) * Multiple 3D images |
Multiple subjects * Single 3D image |
Users can ingest 4D data using either: - The Pillow library, which supports various formats - Or NumPy arrays as a simple alternative
Image datasets are somewhat multi-modal in that, in order to perform supervised learning on them, they must eventually be paired with a
Dataset.Tabularthat acts as itsLabel.
└── Dataset.Image.from_numpy()
Dataset.Image.from_numpy(
arr4D_or_npyPath
, ingest
, rename_columns
, retype
, description
, name
)
Argument |
Type |
Default |
Description |
|---|---|---|---|
arr4D_or_npyPath |
object / str |
Required |
|
ingest |
bool |
None |
See
Registration.
If left blank, ndarrays
will be ingested and npy
will not. Errors if input
is ndarray and
|
rename_columns |
list[str] |
None |
See Registration |
retype |
np.type / dict(column:np.type) |
None |
See Registration |
description |
str |
None |
See Registration |
name |
str |
None |
See Registration |
└── Dataset.Image.from_folder()
Dataset.Image.from_folder(
folder_path
, ingest
, rename_columns
, retype
, description
, name
)
Argument |
Type |
Default |
Description |
|---|---|---|---|
folder_path |
str |
Required |
Folder of images to be ingested via Pillow. All images must be cropped to the same dimensions ahead of time. |
ingest |
bool |
False |
See Registration |
rename_columns |
list[str] |
None |
See Registration |
retype |
np.type / dict(column:np.type) |
None |
See Registration |
description |
str |
None |
See Registration |
name |
str |
None |
See Registration |
└── Dataset.Image.from_urls()
Dataset.Image.from_urls(
urls
, source_path
, ingest
, rename_columns
, retype
, description
, name
)
Argument |
Type |
Default |
Description |
|---|---|---|---|
urls |
list(str) |
Required |
URLs that point to an image to be ingested via Pillow. All images must be cropped to the same dimensions ahead of time. |
source_path |
str |
None |
Optionally record a shared directory, bucket, or FTP site where images are stored. The backend won’t use this information for anything. |
ingest |
bool |
False |
See Registration |
rename_columns |
list[str] |
None |
See Registration |
retype |
np.type / dict(column:np.type) |
None |
See Registration |
description |
str |
None |
See Registration |
name |
str |
None |
See Registration |
1aii. Fetch
The following methods are exposed to end-users in case they want to inspect the data that they have ingested.
└── Dataset.to_arr()
Argument |
Type |
Default |
Description |
|---|---|---|---|
id |
int |
None |
The identifier of the Dataset of interest |
columns |
list(str) |
None |
If left blank, includes all columns |
samples |
list(int) |
None |
If left blank, includes all samples |
Subclass |
Returns |
|---|---|
Tabular |
ndarray.ndim==2 |
Sequence |
ndarray.ndim==3 |
Image |
ndarray.ndim==4 |
└── Dataset.to_df()
Argument |
Type |
Default |
Description |
|---|---|---|---|
id |
int |
None |
The identifier of the Dataset of interest |
columns |
list(str) |
None |
If left blank, includes all columns |
samples |
list(int) |
None |
If left blank, includes all samples |
Subclass |
Returns |
|---|---|
Tabular |
DataFrame |
Sequence |
list(DataFrame) |
Image |
list(list(DataFrame)) |
└── Dataset.to_pillow()
Argument |
Type |
Default |
Description |
|---|---|---|---|
id |
int |
None |
The identifier of the Dataset of interest |
samples |
list(int) |
None |
If left blank, includes all samples |
Subclass |
Returns |
|---|---|
Image |
list(PIL.Image) |
└── Dataset.get_dtypes()
Argument |
Type |
Default |
Description |
|---|---|---|---|
id |
int |
None |
The identifier of the Dataset of interest |
columns |
list(str) |
None |
If left blank, includes all columns |
Regardless of how the initial Dataset.dtype was formatted [e.g. single np.type / str(np.type) / dict(column=np.type)], this function intentionally returns then dtype of each column within a dict(column=str(np.type) format.
1b. Attributes
These are the fields in the Dataset table
Attribute |
Type |
Description |
|---|---|---|
typ |
CharField |
The Dataset type: Tabular, Sequence, Image |
source_format |
CharField |
The file format (Parquet, CSV, TSV) or in-memory class (DataFrame, ndarray) |
source_path |
CharField |
The path of the original file/ folder |
urls |
JSONField |
A list of URLs as an alternative to file paths/ folders |
columns |
JSONField |
List of str-based names for each column |
dtypes |
JSONField |
The type of each column.
Tabular dtype is saved in
|
shape |
JSONField |
Human-readable dictionary
about the dimensions of the
data e.g.
|
sha256_hexdigest |
CharField |
A hash of the data to determine its uniqueness for versioning. |
memory_MB |
IntegerField |
Size of the dataset in megabytes when loaded into memory |
contains_nan |
BooleanField |
Whether or not the dataset contains any blank cells |
header |
PickleField |
|
is_ingested |
BooleanField |
Quick flag to see if the
data was ingested. Exists
to prevent querying the
|
blob |
BlobField |
The raw bytes of the data
obtained via
|
version |
IntegerField |
The auto-incrementing version number assigned to unique datasets that share name |
2. Feature
Determines the columns that will be used as predictive features during training. Columns is always the last dimension shape[-1] of a dataset.
2a. Methods
└── Feature.from_dataset()
Feature.from_dataset(
dataset_id
, include_columns
, exclude_columns
)
Argument |
Type |
Default |
Description |
|---|---|---|---|
dataset_id |
int |
Required |
|
include_columns |
list(str) |
None |
Specify columns that will be included in the Feature. All columns that are not specified will not be included. |
exclude_columns |
list(str) |
None |
Specify columns that will not be included in the Feature. All columns that are not specified will be included. |
If neither
include_columnsnorexclude_columnsis defined, then all columns will be used.
Both
include_columnsandexclude_columnscannot be used at the same time
Fetch
Theses methods wrap Dataset’s fetch methods:
Method |
Arguments |
Returns |
|---|---|---|
to_arr() |
id:int, columns:list(str)=Feature.c olumns, samples:list(int)=None |
ndarray 2D / 3D / 4D |
to_df() |
id:int, columns:list(str)=Feature.c olumns, samples:list(int)=None |
df / list(df) / list(list(df)) |
get_dtypes() |
id:int, columns:list(str)=Feature.c olumns |
dict(column=str(np.type)) |
2b. Attributes
These are the fields in the Feature table
Attribute |
Type |
Description |
|---|---|---|
columns |
JSONField |
The columns included in this featureset |
columns_excluded |
JSONField |
The columns, if any, in the dataset that were not included |
fitted_featurecoders |
PickleField |
When |
dataset |
ForeignKeyField |
Where these columns came from |
3. Label
Determines the column(s) that will be used as a target during supervised analysis. Do no create a Label if you intend to conduct unsupervised/ self-supervised analysis.
3a. Methods
└── Label.from_dataset()
Label.from_dataset(
dataset_id
, columns
)
Argument |
Type |
Default |
Description |
|---|---|---|---|
dataset_id |
int |
Required |
|
columns |
list(str) |
None |
Specify columns that will be included in the Label. If left blank, defaults to all columns. If more than 1 column is provided, then the data in those columns must be in One-Hot Encoded (OHE) format. |
Fetch
Theses methods wrap Dataset’s fetch methods:
Method |
Arguments |
Returns |
|---|---|---|
to_arr() |
id:int, columns:list(str)=Label.col umns, samples:list(int)=None |
ndarray 2D / 3D / 4D |
to_df() |
id:int, columns:list(str)=Label.col umns, samples:list(int)=None |
df / list(df) / list(list(df)) |
get_dtypes() |
id:int, columns:list(str)=Label.col umns |
dict(column=str(np.type)) |
3b. Attributes
These are the fields in the Feature table
Attribute |
Type |
Description |
|---|---|---|
columns |
JSONField |
The column(s) included in this featureset. |
column_count |
IntegerField |
The number of columns in the Label. Used to determine if it is in validated OHE format or not |
unique_classes |
JSONField |
Records all of the different values found in categorical columns. Not used for continuous columns. |
fitted_labelcoder |
PickleField |
When a |
dataset |
ForeignKeyField |
Where these columns came from |
4. Interpolate
If you don’t have time series data then you do not need interpolation.
If you have continuous columns with missing data in a time series, then interpolation allows you to fill in those blanks mathematically. It does so by fitting a curve to each column. Therefore each column passed to an interpolater must satisfy: np.issubdtype(dtype, np.floating).
Interpolation is the first preprocessor because you need to fill in blanks prior to encoding.
pandas.DataFrame.interpolatehttps://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.DataFrame.interpolate.html
Is utilized due to its ease of use, variety of methods, and support of sparse indices. However, it does not follow the
fit/transformpattern like many of the class-based sklearn preprocessors, so the interpolated training data is concatenated with the evalaution split during the interpolation of evaluation splits.
Below are the default settings if interpolate_kwargs=None that get passed to df.interpolate(). In my experience, method=spline produces the best results. However, if either (a) spline fails to fit to your data, or (b) you know that your pattern is linear - then try method=linear.
interpolate_kwargs = dict(
method = 'spline'
, limit_direction = 'both'
, limit_area = None
, axis = 0
, order = 1
)
Because the sample dimension is different for each Dataset Type, they approach interpolation differently.
Dataset Type |
Approach |
|---|---|
Tabular |
Unlike encoders, there is
no |
Sequence |
Interpolation is ran on each 2D sequence separately |
Image |
Interpolation is ran on each 2D channel separately |
4a. LabelInterpolater
Label is intended for a single column, so only 1 Interpolater will be used during Label.preprocess()
4ai. Methods
└── LabelInterpolater.from_label()
LabelInterpolater.from_label(
label_id
, process_separately
, interpolate_kwargs
)
Argument |
Type |
Default |
Description |
|---|---|---|---|
label_id |
int |
Required |
Points to the
|
process_separately |
bool |
True |
Used to restrict the fit to
the training data, this may
be flipped to |
interpolate_kwargs |
dict |
None |
Gets passed to
|
4aii. Attributes
These are the fields in the LabelInterpolater table
Attribute |
Type |
Description |
|---|---|---|
process_separately |
BooleanField |
Whether or not the training data was interpolated by fitting to the entire dataset or not. Indicator of data leakage. |
interpolate_kwargs |
JSONField |
Gets passed to
|
matching_columns |
JSONField |
The columns that were successfully interpolated |
label |
ForeignKeyField |
The Label that this LabelInterpolater is applied to |
4b. FeatureInterpolater
For multivariate datasets, columns/dtypes may need to be handled differently. So we use column/dtype filters to apply separate transformations. If the first transformation’s filter includes a certain column/dtype, then subsequent filters may not include that column/dtype.
4bi. Methods
└── FeatureInterpolater.from_feature()
FeatureInterpolater.from_feature(
feature_id
, process_separately
, interpolate_kwargs
, dtypes
, columns
, verbose
)
Argument |
Type |
Default |
Description |
|---|---|---|---|
feature_id |
int |
Required |
Points to the
|
process_separately |
bool |
True |
Used to restrict the fit to
the training data, this may
be flipped to |
interpolate_kwargs |
dict |
None |
The
|
dtypes |
list(str) |
None |
The dtypes to include |
columns |
list(str) |
None |
The columns to include. Errors if any of the columns were already included by dtypes. |
verbose |
bool |
True |
If True, messages will be printed about the status of the interpolaters as they attempt to fit on the filtered columns |
4bii. Attributes
These are the fields in the FeatureInterpolater table
Attribute |
Type |
Description |
|---|---|---|
idx |
IntegerField |
Zero-based auto-incrementer that counts the number of FeatureInterpolaters attached to a Feature. |
process_separately |
BooleanField |
Whether or not the training data was interpolated by fitting to the entire dataset or not. Indicator of data leakage. |
interpolate_kwargs |
JSONField |
Gets passed to
|
matching_columns |
JSONField |
The columns that matched the filter |
leftover_columns |
JSONField |
The columns that were not included in the filter |
leftover_dtypes |
JSONField |
The dtypes that were not included in the filter |
original_filter |
JSONField |
|
feature |
ForeignKeyField |
The Feature that this FeatureInterpolater is applied to |
5. Encode
Transform data into numerical format that is close to zero. Reference Encoding for more information.
There are two phases of encoding: 1. fit on train - where the encoder learns about the values of the samples made available to it. Ideally, you only want to fit aka learn from your training split so that you are not leaking information from your validation and test spits into your model! However, categorical encoders are always fit on the entire dataset because they are not prone to leakage and any weights tied to empty OHE
inputs will zero-out. 2. transform each split/fold
Only sklearn.preprocessing methods are officially supported, but we have experimented with
sklearn.feature_extraction.text.CountVectorizer
5a. LabelCoder
Label is intended for a single column, so only 1 LabelCoder will be used during Label.preprocess()
Unfortunately, the name “LabelEncoder” is occupied by
sklearn.preprocessing.LabelEncoder
5ai. Methods
└── LabelCoder.from_label()
LabelCoder.from_label(
label_id
, sklearn_preprocess
)
Argument |
Type |
Default |
Description |
|---|---|---|---|
label_id |
int |
Required |
Points to the
|
sklearn_preprocess |
object |
Required |
An instantiated
|
5aii. Attributes
These are the fields in the LabelCoder table
Attribute |
Type |
Description |
|---|---|---|
only_fit_train |
BooleanField |
Whether or not the encoder was fit on the training data or the entire dataset |
is_categorical |
BooleanField |
If the encoder is meant for categorical data, and therefore automatically fit on the entire dataset |
sklearn_preprocess |
PickleField |
The instantiated sklearn.preprocessing class that was fit |
matching_columns |
JSONField |
The columns that matched the dtype/ column name filters |
encoding_dimension |
CharField |
Did the encoder succeed on 1D/ 2D uni-column/ 2D multi-column? |
label |
ForeignKeyField |
The Label that this LabelCoder is applied to |
5b. FeatureCoder
For multivariate datasets, columns/dtypes may need to be handled differently. So we use column/dtype filters to apply separate transformations. If the first transformation’s filter includes a certain column/dtype, then subsequent filters may not include that column/dtype.
5bi. Methods
└── FeatureCoder.from_feature()
FeatureCoder.from_feature(
feature_id
, sklearn_preprocess
, include
, dtypes
, columns
, verbose
)
Argument |
Type |
Default |
Description |
|---|---|---|---|
feature_id |
int |
Required |
Points to the
|
sklearn_preprocess |
object |
Required |
An instantiated
|
include |
bool |
True |
Whether to include or
exclude the dtypes/columns
that match the filter. You
can create a filter for all
columns by setting
|
dtypes |
list(str) |
None |
The dtypes to filter |
columns |
list(str) |
None |
The columns to filter. Errors if any of the columns were already used by dtypes. |
verbose |
bool |
True |
If True, messages will be printed about the status of the encoders as they attempt to fit on the filtered columns |
5bii. Attributes
These are the fields in the FeatureCoder table
Attribute |
Type |
Description |
|---|---|---|
idx |
IntegerField |
Zero-based auto-incrementer that counts the number of FeatureCoders attached to a Feature. |
sklearn_preprocess |
PickleField |
The instantiated sklearn.preprocessing class that was fit |
encoded_column_names |
JSONField |
After the columns are
encoded, what are their
names? OHE appends
|
matching_columns |
JSONField |
The columns that matched the filter |
leftover_columns |
JSONField |
The columns that were not included in the filter |
leftover_dtypes |
JSONField |
The dtypes that were not included in the filter |
original_filter |
JSONField |
|
encoding_dimension |
CharField |
Did the encoder succeed on 1D/ 2D uni-column/ 2D multi-column? |
only_fit_train |
BooleanField |
Whether or not the encoder was fit on the training data or the entire dataset |
is_categorical |
BooleanField |
If the encoder is meant for categorical data, and therefore automatically fit on the entire dataset |
feature |
ForeignKeyField |
The Feature that this FeatureCoder is applied to |
6. Shape
Changes the shape of data. Only supports Features, not Labels.
Reshaping is applied at the end of Feature.preprocess(). So if the feature data has been altered via time series windowing or One Hot Encoder, then those changes will be reflected in the shape that is fed to `
When working with architectures that are highly dimensional such convolutional and recurrent networks (Conv1D, Conv2D, Conv3D / ConvLSTM1D, ConvLSTM2D, ConvLSTM3D), you’ll often find yourself needing to reshape data to fit a layer’s required input shape.
Reducing unused dimensions - When working with grayscale images (1 channel, 25 rows, 25 columns) it’s better to use Conv1D instead of Conv2D.
Adding wrapper dimensions - Perhaps your data is a fit for ConvLSTM1D, but that layer is only supported in the nightly TensorFlow build so you want to add a wrapper dimension in order to use the production-ready ConvLSTM2D.
AIQC favors a “channels_first” (samples, channels, rows, columns) approach as opposed to “channels_last” (samples, rows, columns, channels).
Can’t I just reshape the tensors during the training loop? You could. However, AIQC systemtically provides the shape of features and labels to
Algorith.fn_buildto make designing the topology easier, so it’s best to get the shape right beforehand. Additionally, if you reshape your data within the training loop, then you may also need to reshape the output ofAlgorithm.fn_predictso that it is correctly formatted for automatic post-processing. It’s also more computationally efficient to do the reshaping once up front.
The reshape_indices argument is ultimately fed to np.reshape(newshape). We use index n to point to the value at ndarray.shape[n].
Reshaping by Index
Let’s say we have a 4D feature consisting of 3D images (samples * channels * rows * columns). Our problems is that the images are B&W, so we don’t want a color channel because it would add unecessary dimensionality to our model. So we want to drop the dimension at the shape index 1.
reshape_indices = (0,2,3)
Thus we have wrangled ourselves a 3D feature consisting of 2D images (samples * rows * columns).
Reshaping Explicitly
But what if the dimensions we want cannot be expressed by rearranging the existing indices? If you define a number as a str, then that number will be used as directly as the value at that position.
So if I wanted to add an extra wrapper dimension to my data to serve as a single color channel, I would simply do:
reshape_indices = (0,'1',1,2)
Then couldn’t I just hardcode my shapes with strings? Yes, but
FeatureShaperis applied to all of the splits, which are assumed to have different shapes, which is why we use the indices.
Multiplicative Reshaping
Sometimes you need to stack/nest dimensions. This requires multiplying one shape index by another.
For example, if I have a 3 separate hours worth of data and I want to treat it as 180 minutes, then I need to go from a shape of (3 hours * 60 minutes) to (180 minutes). Just provide the shape indices that you want to multiply in a tuple like so:
<!> if your model is unsupervised (aka generative or self-supervised), then it must output data in “column (aka width) last” shape. Otherwise, automated column decoding will be applied along the wrong dimension.
6a. Methods
└── FeatureShaper.from_feature()
FeatureShaper.from_feature(
feature_id
, reshape_indices
)
Argument |
Type |
Default |
Description |
|---|---|---|---|
feature_id |
int |
Required |
The |
reshape_indices |
tuple(int/str/tuple) |
Required |
See Strategies. |
6b. Attributes
These are the fields of the FeatureShaper table
Attribute |
Type |
Description |
|---|---|---|
reshape_indices |
PickleField |
See #Reshaping-by-Index.Pickle because tuple has no JSON equivalent. |
column_position |
IntegerField |
The shape index used for columns aka width. |
feature |
ForeignKeyField |
The Feature that reshaping is applied to. |
7. Window
Window facilitates sliding windows for a time series Feature. It does not apply to Labels. This is used for unsupervised (aka self-supervised) walk-forward forecasting for time series data.
size_window determine how many timepoints are included in a window, and size_shift determines how many timepoints to slide over before defining a new window.
For example, if we want to be able to predict the next 7 days worth of weather using the past 21 days of weather, then our
size_window=21and oursize_shift=7.
Challenges
Dealing with stratified windowed data demands a systematic approach.
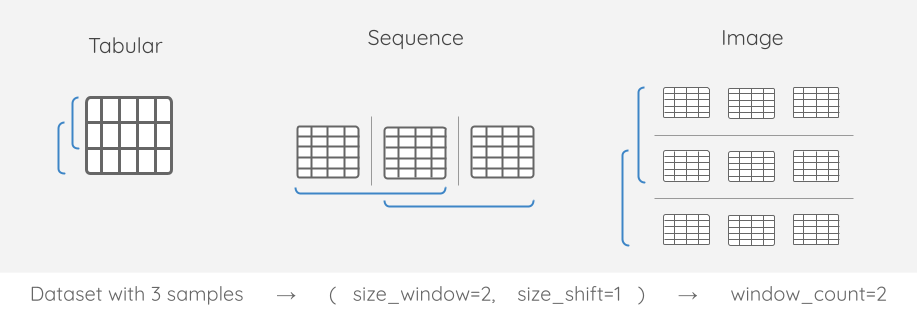
Windowing always increases dimensionality
After data is windowed, its dimensionality increases by 1. Why? Well, originally we had a single time series. However, if we window that data, then we have many time series subsets.
As the highest dimension, it becomes the “sample”
No matter what dimensionality the original data has, it will be windowed along the first dimension.
This means that the windows now serve as the samples, which is important for stratification. If we have a year’s worth of windows, we don’t want all of our training windows to come from the same season. Therefore, Window must be created prior to Splitset.
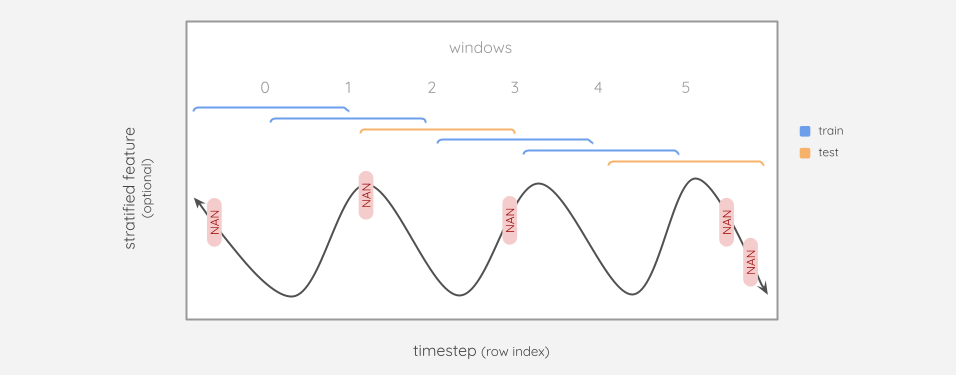
Windowing may causes overlap in splits
In addition to increasing the dimensionality of our data, it makes it harder to nail down the boundaries of our splits in order to prevent data leakage.
As seen in the diagram above, the timesteps of the train and test splits may overlap. So if we are fitting an interpolater to our training split, the first 3 NaNs would be included, but the last 2 would not.
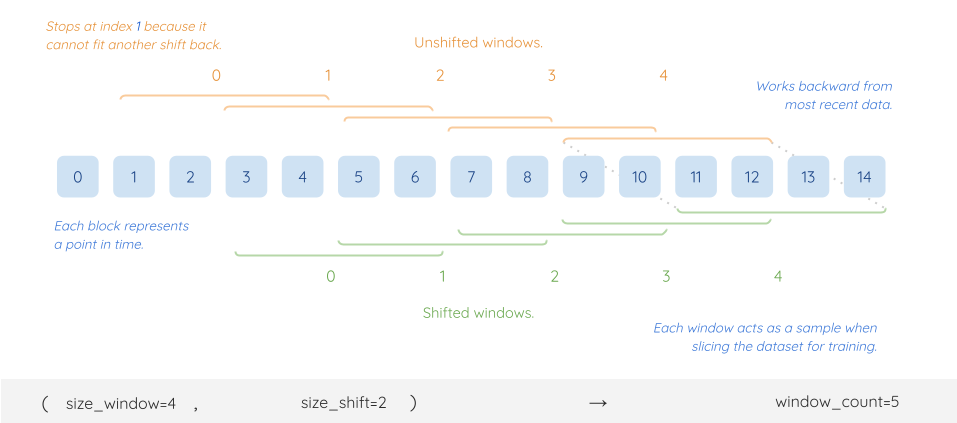
Shifted and unshifted windows
In a walk-forward analysis, we learn about the future by looking at the past. So we need 2 sets of windows:
Unshifted windows (orange in diagram above): represent the past and serves as the features we learn from
Shifted windows (green in diagram above): represent the future and serves as the target we predict
However, when conducting inference, we are trying to predict the shifted windows not learn from them. So we don’t need to record any shifted windows.
7a. Methods
└── Window.from_feature()
Window.from_feature(
feature_id
, size_window
, size_shift
, record_shifted
)
Argument |
Type |
Default |
Description |
|---|---|---|---|
dataset_id |
int |
Required |
|
size_window |
int |
Required |
The number of timesteps to include in a window. |
size_shift |
int |
Required |
The number of timesteps to shift forward. |
record_shifted |
bool |
True |
Whether or not we want to keep a shifted set of windows around. During pure inference, this is False. |
7b. Attributes
These are the fields of the Window table
Attribute |
Type |
Description |
|---|---|---|
size_window |
IntegerField |
Number of timesteps in each window |
size_shift |
IntegerField |
The number of timesteps in the shift forward. |
window_count |
IntegerField |
Not a relationship count! Number of windows in the dataset. This becomes the new samples dimension for stratification. |
samples_unshifted |
JSONField |
Underlying sample indices of each window in the past-shifted windows. |
samples_shifted |
JSONField |
Underlying sample indices of each window in the future-shifted windows. |
feature |
ForeignKeyField |
The Feature that this windowing is applied to |
8. Splitset
Used for sample stratification. Reference Stratification section of the Explainer.
Split |
Description |
|---|---|
train |
The samples that the model will be trained upon. Later, we’ll see how we can make cross-folds from our training split. Unsupervised learning will only have a training split. |
validation (optional) |
The samples used for training evaluation. Ensures that the test set is not revealed to the model during training. |
test (optional) |
The samples the model has never seen during training. Used to assess how well the model will perform on unobserved, natural data when it is applied in the real world aka how generalizable it is. |
Because Splitset groups together all of the data wrangling entities (Features, Label, Folds) it essentially represents a Pipeline, which is why it bears the name Pipeline in the High-Level API.
Cross-Validation
Cross-validation is triggered by fold_count:int during Splitset creation. Reference the scikit-learn documentation to learn more about cross-validation.
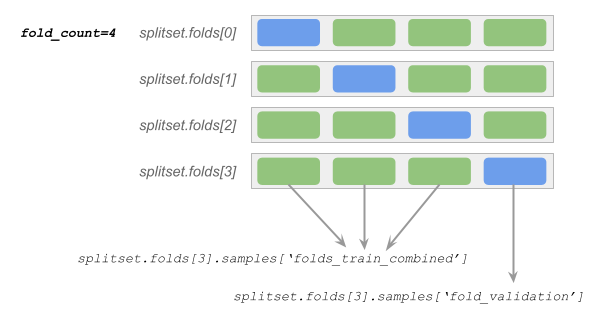
Each row in the diagram above is a Fold object.
Each green/blue box represents a bin of stratified samples. During preprocessing and training, we rotate which blue bin serves as the validation samples (fold_validation). The remaining green bins in the row serve as the training samples (folds_train_combined).
Let’s say we defined fold_count=5. What are the implications?
Creates 5
Foldsrelated to aSplitset.5x more preprocessing and caching; each
fold_validationis excluded from thefitonfolds_train_combinared. Fits are saved to theorm.Foldobject as opposed to theorm.Feature/Labelobjects.5x more models will be trained for each experiment.
5x more evaluation.
Disclaimer about inherent limitations & challenges
Do not use cross-validation unless the distribution of each resulting fold (total sample count divided by fold_count) is representatitve of your broader sample population. If you are ignoring that advice and stretching to perform cross-validation, then at least ensure that the total sample count is evenly divisble by fold_count. Both of these tips help avoid poorly stratified/ undersized folds that seem to perform either unjustifiably well (100% accuracy when only the most common label class is present) or poorly (1 incorrect prediction in a small fold negatively skews an otherwise good model).
If you’ve ever performed cross-validation manually with too few samples, then you’ll know that it’s easy enough to construct the folds, but then it’s a pain to calculate performance metrics (e.g.
zero_division, absent OHE classes) due to the absence of outlying classes and bins. Time has been invested to handle these scenarios elegantly so that folds can be treated as first-class-citizens alongside splits. That being said, if you try to do something undersized like multi-label classification using 150 samples then you may run into errors during evaluation.
Samples Cache
Each Splitset has as cache_path attribute, which represents a local directory where preprocessed data is stored during training & evaluation.
The output of feature.preprocess() and label.preprocess() are written to this folder prior to training so that:
Each Job does not have to preprocess data from scratch.
The original data does not need to be held in memory between Jobs.
└── aiqc/cache/samples/splitset_uid
└── <fold_index> | "no_fold"
└── <split>
└── label.npy
└── feature_<i>.npy
<fold_index>is a folder for each Fold, since they have different samples. Whereas “no_fold” is a single folder for a regular splitset where there are no folds. ‘no_fold’ just keeps the folder depth uniform for regular splitsets<split>: Thesamples[<split>]of interest: ‘train’, ‘validation’, ‘folds_train_combined’, ‘fold_validation’, ‘test’.feature_<n>accounts for Splitsets with more than 1 Feature.
The Splitset.cache_hot:bool argument indicates whether or not the cache for that splitset is populated or not.
Samples are automatically cached during Queue.run_jobs().
See also: orm.splitset.clear_cache() and utils.config.clear_cache_all()
8a. Methods
└── Splitset.make()
Splitset.make(
feature_ids
, label_id
, size_test
, size_validation
, bin_count
, fold_count
, unsupervised_stratify_col
, name
, description
, predictor_id
)
Argument |
Type |
Default |
Description |
|---|---|---|---|
feature_ids |
list(int) |
Required |
Multiple |
label_id |
int |
None |
The Label to be used as a target for supervised analysis. Must have the sample number of samples as the Features. |
size_test |
float |
None |
Percent of samples to be
placed into the test split.
Must be |
size_validation |
float |
None |
Percent of samples to be
placed into the validation
split. Must be |
bin_count |
int |
None |
For continous stratification columns, how many bins (aka quantiles) should be used? |
fold_count |
int |
None |
The number or cross-validation folds to generate. See Cross-Validation. |
unsupervised_stratify_c ol |
str |
None |
Used during unsupervised analysis. Specify a column from the first Feature in feature_ids to use for stratification. For example, when forecasting, it may make sense to stratify by the day of the year. |
name |
str |
None |
Used for versioning a pipeline (collection of inputs, label, and stratification). Two versions cannot have identical attributes. |
description |
str |
None |
What is unique about this this pipeline? |
size_train = 1.00 - (size_test + size_validation)the backend ensures that the sizes sum to 1.00
How does continuous binning work? Reference the handy
Pandas.qcut()and the source codepd.qcut(x=array_to_bin, q=bin_count, labels=False, duplicates='drop')for more detail.
└── Splitset.cache_samples()
See Samples Cache section for a description
Splitset.cache_samples(id)
Argument |
Type |
Default |
Description |
|---|---|---|---|
id |
int |
Required |
The identifier of the Splitset of interest |
└── Splitset.clear_cache()
See Samples Cache section for a description. Deletes the entire directory located at Splitset.cache_path.
Splitset.clear_cache(id)
Argument |
Type |
Default |
Description |
|---|---|---|---|
id |
int |
Required |
The identifier of the Splitset of interest |
└── Splitset.fetch_cache()
See Samples Cache section for a description. This fetches a specific file from the cache.
Splitset.fetch_cache(
id
, split
, label_features
, fold_id
, library
)
Argument |
Type |
Default |
Description |
|---|---|---|---|
id |
int |
Required |
The identifier of the Splitset of interest |
split |
int |
Required |
The |
label_features |
str |
Required |
Either |
fold_id |
int |
None |
The identifier of the Fold of interest, if any |
library |
str |
None |
If |
|
Returns |
|---|---|
|
|
|
|
8b. Attributes
These are the fields of the Splitset table
Attribute |
Type |
Description |
|---|---|---|
cache_path |
CharField |
Where the splitset stores its cached samples |
cache_hot |
BooleanField |
If the samples are currently stored in the cache |
samples |
JSONField |
The bins that splits have
been stratified into
|
sizes |
JSONField |
Human-readable sizes of the
splits
|
supervision |
CharField |
Either “supervised” or “unsupervised” if the Splitset has a Label. |
has_validation |
BooleanField |
Logical flag indicating if this Splitset has a validation split. |
fold_count |
IntegerField |
The number of
cross-validation |
bin_count |
IntegerField |
The number of bins used to stratify a continuous column label or unsupervised_stratify column |
**unsupervised_stratifyCol ** |
CharField |
Used during unsupervised analysis. Specify a column from the first Feature in feature_ids to use for stratification. For example, when forecasting, it may make sense to stratify by the day of the year. |
key_train |
CharField |
|
key_evaluation |
CharField |
|
key_test |
CharField |
|
version |
IntegerField |
[TBD] |
label |
ForeignKeyField |
The Label, if any, that supervises this splitset |
predictor |
DeferredForeignKey |
During inference, a new
Splitset of samples to be
predicted may attach to a
Predictor. Samples dict
will bear the key of the
|
These are the fields of the Fold table
Attribute |
Type |
Description |
|---|---|---|
idx |
IntegerField |
Zero-based auto-incrementer that counts the Folds |
samples |
JSONField |
Contains the sample indices
of the training folds and
leftout validation fold, as
well as any validation and
test splits defined in the
regular Splitset.samples
|
fitted_labelcoder |
PickleField |
When |
fitted_featurecoders |
PickleField |
When |
splitset |
ForeignKeyField |
The Splitset that this Fold belongs to |
9. Algorithm
Now that our data has been prepared, we transition to the 2nd half of the ORM where the focus is the logic that will be applied to that data.
The Algorithm contains all of the components needed to construct, train, and use our model.
Reference the tutorials for examples of how Algorithms are defined.
PyTorch Fit
Provides an abstraction that eliminates the boilerplate code normally required to train and evaluate a PyTorch model.
Before training - it shuffles samples, batches samples, and then shuffles batches.
During training - it calculates batch loss, epoch loss, and epoch history metrics.
After training - it calculates metrics for each split.
model, history = utils.pytorch.fit(
# These arguments come directly from `fn_train`
model
, loser
, optimizer
, train_features
, train_label
, eval_features
, eval_label
# These arguments are user-defined
, epochs
, batch_size
, enforce_sameSize
, allow_singleSample
, metrics
)
User-Defined Arguments |
Type |
Default |
Description |
|---|---|---|---|
epochs |
int |
30 |
The number of times to loop over the features |
batch_size |
int |
5 |
Divides features and lables into chunks to be trained upon |
enforce_sameSize |
bool |
True |
If |
allow_singleSample |
bool |
False |
If |
metrics |
list(torchmetrics.metric()) |
None |
List of instantiated
|
History Metrics
The goal of the Predictor.history object is to record the training and evaluation metrics at the end of each epic so that they can be interpretted in the learning curve plots. Reference the evaluation section.
Keras: any
metrics=[]specified are automatically added to theHistorycallback object.PyTorch: if you use
fitseen above, then you don’t need to worry about this. Users are responsible for calculating their own metrics (we recommend thetorchmetricspackage) and placing them into ahistorydictionary that mirrors the schema of the Keras history object. Reference the torch examples.
The schema of the
historydictionary is as follows:dict(<metric>:ndarray, val_<metric>=ndarray). For example, if you wanted to record the history of the ‘loss’ and ‘accuracy’ metrics manually for PyTorch, you would construct it like so:
history = dict(
loss = ndarray
, val_loss = ndarray
, accuracy = ndarray
, val_accuracy = ndarray
)
TensorFlow Early Stopping
Early stopping isn’t just about efficiency in reducing the number of epochs. If you’ve specified 300 epochs, there’s a chance your model catches on to the underlying patterns early, say around 75-125 epochs. At this point, there’s also good chance what it learns in the remaining epochs will cause it to overfit on patterns that are specific to the training data, and thereby and lose it’s simplicity/ generalizability.
The
metric=val_*prefix refers to the evaluation samples.Remember, regression does not have accuracy metrics.
TrainingCallback.MetricCutoffis a custom class we wrote to make early stopping easier, so you won’t find information about it in the official Keras documentation.
Placed within fn_train:
from aiqc.utils.tensorflow import TrainingCallback
#Define one or more metrics to monitor.
metrics_cuttoffs = [
dict(metric='accuracy', cutoff=0.96, above_or_below='above'),
dict(metric='loss', cutoff=0.1, above_or_below='below')
dict(metric='val_accuracy', cutoff=0.96, above_or_below='above'),
dict(metric='val_loss', cutoff=0.1, above_or_below='below')
]
cutoffs = TrainingCallback.MetricCutoff(metrics_cuttoffs)
# Pass it into keras callbacks
model.fit(
# other fit args
callbacks = [cutoffs]
)
Tip: try using a
val_accuracythreshold by itself for best results
9a. Methods
Assemble an architecture consisting of components defined in functions.
The **hp kwargs are common to every Algorithm function except fn_predict. They are used to systematically pass a dictionary of hyperparameters into these functions. See Hyperparameters.
└── Algorithm.make()
Algorithm.make(
library
, analysis_type
, fn_build
, fn_train
, fn_predict
, fn_lose
, fn_optimize
)
Argument |
Type |
Default |
Description |
|---|---|---|---|
library |
str |
Required |
‘keras’ or ‘pytorch’
depending on the type of
model defined in
|
analysis_type |
str |
Required |
‘classification_binary’, ‘classification_multi’, or ‘regression’. Unsupervised/ self-supervised falls under regression. Used to determine which performance metrics are run. Errors if it is incompatible with the Label provided: e.g. classification_binary is incompatible with an np.floating Label.column. |
fn_build |
func |
Required |
See below. Build the model architecture. |
fn_train |
func |
Required |
See below. Train the model. |
fn_predict |
func |
None |
See below. Run the model. |
fn_lose |
func |
None |
See below. Calculate loss. |
fn_optimize |
func |
None |
See below. Optimization strategy. |
Required Functions
def fn_build(
features_shape:tuple
, label_shape:tuple
, **hp:dict
):
# Define tf/torch model
return model
The
*_shapearguments contain the shape of a single sample, as opposed to a batch or entire dataset.features_shapeis plural because it may contain the shape of multiple features. However, if only 1 feature was used then it will not be inside a list.
def fn_train(
model:object
, loser:object
, optimizer:object
, train_features:ndarray
, train_label:ndarray
, eval_features:ndarray
, eval_label:ndarray
, **hp:dict
):
# Define training/ eval loop.
# See `utils.pytorch.fit`
# if tensorflow
return model
# if torch
# See `utils.pytorch.fit` and history metrics below
return history:dict, model
Optional Functions
Where are the defaults for optional functions defined? See utils.tensorflow and utils.pytorch for examples of loss, optimization, and prediction.
def fn_predict(model:object, features:ndarray):
#if classify. predictions always ordinal, never OHE.
return prediction, probabilities #both as ndarray
#if regression
return prediction #ndarray
def fn_lose(**hp:dict):
# Define tf/torch loss function
return loser
def fn_optimize(**hp:dict):
# Define tf/torch optimizer
return optimizer
└── Algorithm.get_code()
Returns the strings of the Algorithm functions:
dict(
fn_build = aiqc.utils.dill.reveal_code(Algorithm.fn_build)
, fn_lose = aiqc.utils.dill.reveal_code(Algorithm.fn_lose)
, fn_optimize = aiqc.utils.dill.reveal_code(Algorithm.fn_optimize)
, fn_train = aiqc.utils.dill.reveal_code(Algorithm.fn_train)
, fn_predict = aiqc.utils.dill.reveal_code(Algorithm.fn_predict)
)
9b. Attributes
These are the fields of the Algorithm table
Attribute |
Type |
|---|---|
library |
CharField |
analysis_type |
CharField |
fn_build |
BlobField |
fn_lose |
BlobField |
fn_optimize |
BlobField |
fn_train |
BlobField |
fn_predict |
BlobField |
See #9.-Algorithm for descriptions
10. Hyperparameters
As mentioned in Algorithm, the **hp argument is used to systematically pass hyperparameters into the Algorithm functions.
For example, given the follow set of hyperparamets:
hyperparameters = dict(
epoch_count = [30]
, learning_rate = [0.01]
, neuron_count = [24, 48]
)
A grid search would produce the 2 unique Hyperparamcombo’s:
[
dict(
epoch_count = 30
, learning_rate = 0.01
, neuron_count = 24 #<-- varies
)
, dict(
epoch_count = 30
, learning_rate = 0.01
, neuron_count = 48 #<-- varies
)
]
We access the current value in our model functions like so: hp['neuron_count'].
10a. Methods
└── Hyperparamset.from_algorithm()
Hyperparamset.from_algorithm(
algorithm_id
, hyperparameters
, search_count
, search_percent
)
Argument |
Type |
Default |
Description |
|---|---|---|---|
algorithm_id |
int |
Required |
The |
hyperparameters |
dict(str:list) |
Required |
See example in Hyperparameters. Must be JSON compatible. |
search_count |
int |
None |
Randomly select n hyperparameter combinations to test. Must be greater than 1. No upper limit, it will test all combinations if number of combinations is exceeded. |
search_percent |
float |
None |
Given all of the available
hyperparameter
combinations, search x%.
Between |
“Bayesian TPE (Tree-structured Parzen Estimator)” via
hyperopthas been suggested as a future area to explore, but it does not exist right now.
10b. Attributes
These are the fields of the Hyperparamset table
Attribute |
Type |
Description |
|---|---|---|
hyperparameters |
JSONField |
The original
|
search_count |
IntegerField |
The number of randomly selected combinations of hyperparameters |
search_percent |
FloatField |
The percent of randomly selected combinations of hyperparameters |
algorithm |
ForeignKeyField |
The |
These are the fields of the Hyperparamcombo table
Attribute |
Type |
Description |
|---|---|---|
idx |
IntegerField |
Zero-based counts the number of the number of hyperparamcombos |
hyperparameters |
JSONField |
The specific combination of hyperparameters that will be fed to the Algorithm functions |
hyperparamset |
ForeignKeyField |
The Hyperparamset that this combination of hyperparameters was derived from |
11. Queue
The Queue is the central object of the “logic side” of the ORM. It ties together everything we need to run training Job’s for hyperparameter tuning. That’s why it is referred to as an Experiment in the High-Level API.
11a. Methods
└── Queue.from_algorithm()
Queue.from_algorithm(
algorithm_id
, splitset_id
, repeat_count
, permute_count
, hyperparamset_id
, description
)
Argument |
Type |
Default |
Description |
|---|---|---|---|
algorithm_id |
int |
Required |
The |
splitset_id |
int |
Required |
The |
repeat_count |
int |
1 |
Each job will be repeat n
times. Designed for use
with random weight
initialization (aka
non-deterministic). This is
why 1 |
permute_count |
int |
3 |
Triggers a shuffled
permutation of each
training data column to
determine which columns
have the most impact on
loss in comparison baseline
training loss:
|
hyperparamset_id |
int |
None |
The |
description |
str |
None |
What is unique about this experiment? |
└── Queue.run_jobs()
Jobs are simply ran on a loop on the main process.
Stop the queue with a keyboard interrupt e.g. ctrl+Z/D/C in Python shell or i,i in Jupyter. It is listening for interupts so it will usually stop gracefully. Even if it errors upon during interrupt, it’s not a problem. You can rerun the queue and it will resume on the same job it was running previously.
Queue.run_jobs(id)
Argument |
Type |
Default |
Description |
|---|---|---|---|
id |
int |
Required |
The identifier of the Queue of interest |
└── Queue.plot_performance()
Plots every model trained by the queue for comparison.
X axis = loss
Y axis = score
Queue.plot_performance(
id
, call_display
, max_loss
, min_score
, score_type
, height
)
Argument |
Type |
Default |
Description |
|---|---|---|---|
id |
int |
Required |
The identifier of the Queue of interest |
call_display |
bool |
True |
If |
max_loss |
float |
None |
Models with any split with higher loss than this threshold will not be plotted. |
min_score |
typ |
None |
Models with any split with a lower score than this threshold will not be plotted. |
score_type |
typ |
None |
Defaults to |
height |
typ |
None |
Default height is |
└── Queue.metrics_df()
Displays metrics for every split/fold of every model.
Queue.metrics_df(
id
, selected_metrics
, sort_by
, ascending
)
Argument |
Type |
Default |
Description |
|---|---|---|---|
id |
int |
Required |
The identifier of the Queue of interest |
ascending |
typ |
False |
Descending if False. |
selected_metrics |
list(str) |
None |
If you get overwhelmed by the variety of metrics returned, then you can include the ones you want selectively by name. |
sort_by |
str |
None |
You can sort the dataframe by any column name. |
└── Queue.metricsAggregate_df()
Aggregate statistics about every metric of every model trained in the Queue – displays the average, median, standard deviation, minimum, and maximum across all splits/folds.
Queue.metricsAggregate_df(
id
, ascending = False
, selected_metrics = None
, selected_stats = None
, sort_by = None
)
Argument |
Type |
Default |
Description |
|---|---|---|---|
id |
int |
Required |
The identifier of the Queue of interest |
ascending |
typ |
False |
Descending if False. |
selected_metrics |
list(str) |
None |
If you get overwhelmed by the variety of metrics returned, then you can include the ones you want selectively by name. |
sort_by |
str |
None |
You can sort the dataframe by any column name. |
11b. Attributes
These are the fields of the Queue table
Attribute |
Type |
Description |
|---|---|---|
repeat_count |
IntegerField |
The number of times to repeat each Job. |
total_runs |
IntegerField |
The total number of models to be trained as a result of this queue being created. |
permute_count |
IntegerField |
Number of permutations to run on each column before taking the median impact on loss. 0 means permutation was skipped. |
runs_completed |
IntegerField |
Counts the runs that have actually finished |
algorithm |
ForeignKeyField |
The model functions to use during training and evaluation |
splitset |
ForeignKeyField |
The pipeline of samples to feed to the models during training and evaluation |
hyperparamset |
ForeignKeyField |
Contains all of the hyperparameters to be used for the Jobs |
12. Job
The Queue spawns Job’s. A Job is like a spec/ manifest for training a model. It may be repeated.
# jobs = Hyperamset.hyperamcombo.count() * Queue.repeat_count * splitset.folds.count()
12a. Methods
There are no noteworthy, user-facing methods for the Job class
12b. Attributes
These are the fields of the Job table
Attribute |
Type |
Description |
|---|---|---|
repeat_count |
IntegerField |
The number of times this Job is to be repeated |
queue |
ForeignKeyField |
The Queue this Job was created by |
hyperparamcombo |
ForeignKeyField |
The parameters this Job uses |
fold |
ForeignKeyField |
The cross-validation samples that this Job uses |
13. Predictor
As the Jobs finish, they save the model and history metrics within a Predictor object.
13a. Methods
└── Predictor.get_model()
predictor.get_model(id)
Handles fetching and initializing the model (and PyTorch optimizer) from Predictor.model_file and Predictor.input_shapes
Argument |
Type |
Default |
Description |
|---|---|---|---|
id |
int |
None |
The identifier of the Predictor of interest |
└── Predictor.get_hyperparameters()
This is a shortcut to fetch the hyperparameters used to train this specific model. as_pandas toggles between dict() and DataFrame.
Predictor.get_hyperparameters(id, as_pandas)
Argument |
Type |
Default |
Description |
|---|---|---|---|
id |
int |
None |
The identifier of the Predictor of interest |
as_pandas |
bool |
True |
If |
└── Predictor.plot_learning_curve()
A learning curve will be generated for each train-evaluation pair of metrics in the Predictor.history dictionary
Predictor.plot_learning_curve(
id
, skip_head
, call_display
)
Argument |
Type |
Default |
Description |
|---|---|---|---|
id |
int |
None |
The identifier of the Predictor of interest |
skip_head |
bool |
True |
Skips displaying the first 15% of epochs. Loss values in the first few epochs can often be extremely high before they plummet and become more gradual. This really stretches out the graph and makes it hard to see if the evaluation set is diverging or not. |
call_display |
bool |
True |
If |
13b. Attributes
These are the fields of the Predictor table
Attribute |
Type |
Description |
|---|---|---|
repeat_index |
IntegerField |
Counts how many predictors have been trained using a Job spec |
time_started |
DateTimeField |
When the Job started |
time_succeeded |
DateTimeField |
When the Job finished |
time_duration |
IntegerField |
Total time in seconds it took to complete the Job |
model_file |
BlobField |
Contains a dilled (advanced
Pickle) of the trained
model. See
|
features_shapes |
PickleField |
tuple or list of tuples containing the np.shape(s) of feature(s) |
label_shape |
PickleField |
tuple containing np.shape of a single sample’s label |
history |
JSONField |
Contains the training history loss/metrics |
is_starred |
BooleanField |
Flag denoting if this model is of interest |
job |
ForeignKeyField |
The Job that trained this Predictor |
14. Prediction
When data is fed through a Predictor, you get a Prediction. During training, Predictions are automatically generated for every split/fold in the Queue.splitset.
14a. Methods
└── Prediction.calc_featureImportance()
This method is provided for conducting feature importance after training. It was decoupled from training for the following reasons:
Permutation is computationally expensive, especially for many-columned datasets.
We don’t care about the feature importance of our best models.
What data is used when calculating feature importance? All splits/folds are concatenated back into a single dataset. This assumes that all splits/folds are relatively equally balanced with respect to their label values. For example, if you have unbalanced multi-labels (55:35:10 distribution of classes) then a given feature’s importance may be biased based on how well it predicts the larger class. For binary classification scenarios, this should not matter as much since predicting one class also helps in predicting the opposite class.
Upon completion it will update the Prediction.feature_importance and Prediction.permute_count attributes.
Prediction.calc_featureImportance(id, permute_count)
Argument |
Type |
Default |
Description |
|---|---|---|---|
id |
int |
None |
The identifier of the Prediction of interest |
permute_count |
int |
Required |
The count determines how
many times the shuffled
permutation is ran before
taking the median loss.
|
└── Prediction.importance_df()
Returns a dataframe of feature columns ranked by their median importance
Prediction.importance_df(id, top_n, feature_id)
Argument |
Type |
Default |
Description |
|---|---|---|---|
id |
int |
None |
The identifier of the Prediction of interest |
top_n |
int |
None |
The number of columns to return |
feature_id |
int |
None |
Limit returned columns to a specific feature identifier |
└── Prediction.plot_feature_importance()
Plots prediction.feature_importance if Queue.permute_count>0 or Prediction.calc_featureImportance() was ran after the fact.
Prediction.plot_feature_importance(
id
, call_display
, top_n
, height
, margin_left
)
Argument |
Type |
Default |
Description |
|---|---|---|---|
id |
int |
None |
The identifier of the Prediction of interest |
call_display |
bool |
True |
If |
top_n |
int |
10 |
The number of features to display. If greater than the actual number of features, it just returns all features. |
boxpoints |
object |
False |
Determines how whiskers,
outliers, and points are
shown. Options are:
|
height |
int |
None |
If |
margin_left |
int |
None |
If |
└── Prediction.plot_roc_curve()
Receiver operating curve (ROC) for classification metrics.
Prediction.plot_roc_curve(id, call_display)
Argument |
Type |
Default |
Description |
|---|---|---|---|
id |
int |
None |
The identifier of the Prediction of interest |
call_display |
bool |
True |
If |
└── Prediction.plot_precision_recall()
Precision/recall curve for classification metrics.
Prediction.plot_precision_recall(id, call_display)
Argument |
Type |
Default |
Description |
|---|---|---|---|
id |
int |
None |
The identifier of the Prediction of interest |
call_display |
bool |
True |
If |
└── Prediction.plot_confusion_matrix()
Confusion matrices for classification metrics.
Prediction.plot_confusion_matrix(id, call_display)
Argument |
Type |
Default |
Description |
|---|---|---|---|
id |
int |
None |
The identifier of the Prediction of interest |
call_display |
bool |
True |
If |
└── Prediction.plot_confidence()
Plot the binary/multi-label classification probabilities for a single sample.
Prediction.plot_confidence(
id,
, prediction_index
, height
, call_display
, split_name
)
Argument |
Type |
Default |
Description |
|---|---|---|---|
id |
int |
None |
The identifier of the Prediction of interest |
prediction_index |
int |
0 |
The index of the sample of interest |
height |
int |
175 |
Force the height of the chart. |
call_display |
bool |
True |
If |
split_name |
int |
None |
The identifier of the Prediction of interest |
14b. Attributes
Attribute |
Type |
Description |
|---|---|---|
predictions |
PickleField |
Decoded predictions ndarray for per split/ fold/ inference |
permute_count |
IntegerField |
The number of times this feature importance permuted each column |
feature_importance |
JSONField |
Importance of each column.
Only calculated for
training split/fold.
Schema:
|
probabilities |
PickleField |
Prediction probabilities
per split/ fold. |
metrics |
PickleField |
Statistics for each split/fold that vary based on the analysis_type. |
metrics_aggregate |
PickleField |
Contains the average, median, standard deviation, minimum, and maximum for each statistic across all splits/folds. |
plot_data |
PickleField |
Metrics reformatted for plot functions. |
Evaluation
To see the visualization of performance metrics of Queue, Predictor and Prediction in action – reference the Evaluation documentation.