TensorFlow: Image Classify Binary
Brain Tumor Detection via Binary Classification of Magnetic Resonance Imaging (MRI) Scans
💾 Data
Reference Example Datasets for more information.
This dataset is comprised of:
Features = folder of magnetic resonance imaging (MRI) of brain samples.
Labels = tabular data denoting the presence of a tumor.
[2]:
from aiqc import datum
df = datum.to_df('brain_tumor.csv')
[4]:
from aiqc.orm import Dataset
dataset_label = Dataset.Tabular.from_df(df)
dataset_label.to_df().head(3)
[4]:
| status | size | count | symmetry | url | |
|---|---|---|---|---|---|
| 0 | 0 | 0 | 0 | None | https://raw.githubusercontent.com/aiqc/aiqc/ma... |
| 1 | 0 | 0 | 0 | None | https://raw.githubusercontent.com/aiqc/aiqc/ma... |
| 2 | 0 | 0 | 0 | None | https://raw.githubusercontent.com/aiqc/aiqc/ma... |
[5]:
urls = datum.get_remote_urls(manifest_name='brain_tumor.csv')
dataset_img = Dataset.Image.from_urls(urls)
🖼️ Ingesting Images 🖼️: 100%|████████████████████████| 80/80 [00:14<00:00, 5.49it/s]
[6]:
images_pillow = dataset_img.to_pillow(samples=[6,47])
[7]:
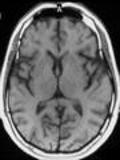
images_pillow[0]
[7]:
[8]:
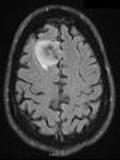
images_pillow[1]
[8]:
🚰 Pipeline
Reference High-Level API Docs for more information.
[10]:
from aiqc.mlops import Pipeline, Input, Target, Stratifier
from sklearn.preprocessing import FunctionTransformer
from aiqc.utils.encoding import div255, mult255
[11]:
pipeline = Pipeline(
Input(
dataset = dataset_img,
encoders = Input.Encoder(FunctionTransformer(div255, inverse_func=mult255)),
reshape_indices = (0,2,3)#reshape for Conv1D grayscale.
),
Target(
dataset = dataset_label,
column = 'status'
),
Stratifier(
size_test = 0.14,
size_validation = 0.22
)
)
🧪 Experiment
Reference High-Level API Docs for more information.
[12]:
from aiqc.mlops import Experiment, Architecture, Trainer
import tensorflow as tf
from tensorflow.keras import layers as l
from aiqc.utils.tensorflow import TrainingCallback
[13]:
def fn_build(features_shape, label_shape, **hp):
m = tf.keras.models.Sequential()
# Convolutional block.
for i in range(hp['blocks']):
# Starts big and gets smaller
multiplier = hp['blocks'] - i
if (i == 0):
m.add(l.Conv1D(
hp['filters']*multiplier
, input_shape=features_shape # <-- the only difference
, kernel_size=hp['kernel_size']
, padding='same'
, kernel_initializer=hp['cnn_init']
))
elif (i > 0):
m.add(l.Conv1D(
hp['filters']*multiplier
, kernel_size=hp['kernel_size']
, padding='same'
, kernel_initializer=hp['cnn_init']
))
m.add(l.Activation('relu'))
m.add(l.MaxPool1D(pool_size=hp['pool_size']))
m.add(l.Dropout(0.4))
m.add(l.Flatten())
# Dense block.
for i in range(hp['dense_blocks']):
m.add(l.Dense(hp['dense_neurons']))
m.add(l.BatchNormalization())
m.add(l.Activation('relu'))
m.add(l.Dropout(0.4))
# Output layer.
m.add(l.Dense(label_shape[0], activation='sigmoid'))
return m
[19]:
def fn_train(
model, loser, optimizer,
train_features, train_label,
eval_features, eval_label,
**hp
):
model.compile(
optimizer = optimizer
, loss = loser
, metrics = ['accuracy']
)
# Early stopping.
metric_cutoffs = [
dict(metric='val_accuracy', cutoff=0.90, above_or_below='above')
, dict(metric='accuracy', cutoff=0.90, above_or_below='above')
, dict(metric='val_loss', cutoff=1.00, above_or_below='below')
, dict(metric='loss', cutoff=1.00, above_or_below='below')
]
cutoffs = TrainingCallback.MetricCutoff(metric_cutoffs)
callbacks = [tf.keras.callbacks.History(), cutoffs]
# Training runs.
model.fit(
train_features, train_label
, validation_data = (eval_features, eval_label)
, verbose = 0
, batch_size = hp['batch_size']
, epochs = hp['epoch_count']
, callbacks = callbacks
)
return model
[20]:
hyperparameters = dict(
blocks = [4]
, filters = [24]
, pool_size = [2]
, kernel_size = [3]
, batch_size = [8] #8 did best. tried 5 and 6 again, no.
, cnn_init = ['normal'] #default is glorot
, dense_blocks = [1]
, dense_neurons = [32] #64 did well.
, epoch_count = [100]
)
[23]:
experiment = Experiment(
Architecture(
library = "keras"
, analysis_type = "classification_binary"
, fn_build = fn_build
, fn_train = fn_train
, hyperparameters = hyperparameters
),
Trainer(
pipeline = pipeline,
repeat_count = 2
)
)
[24]:
experiment.run_jobs()
🔮 Training Models 🔮: 100%|██████████████████████████████████████████| 2/2 [00:20<00:00, 10.47s/it]
📊 Visualization & Interpretation
For more information on visualization of performance metrics, reference the Dashboard documentation.